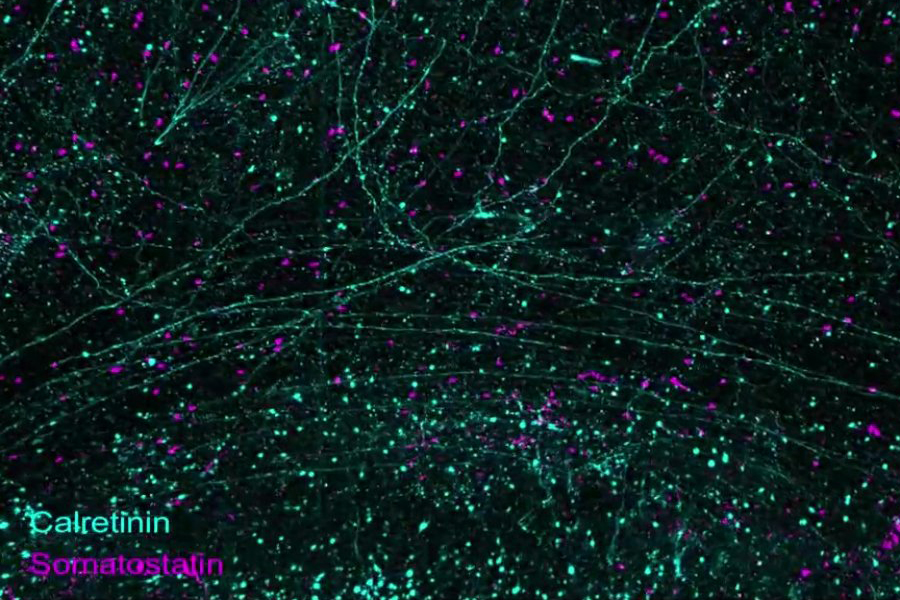
For decades, neuroscientists have aspired to observe and understand the intricate workings of the human brain in its complete form. A groundbreaking study published in Science by a team from MIT showcases a cutting-edge technology pipeline that allows for high-resolution imaging, precise labeling, and detailed processing of full brain hemispheres from two donors – one diagnosed with Alzheimer’s disease and the other healthy.
“Our approach facilitates comprehensive imaging of human brain tissue at various levels, from single synapses to entire brain hemispheres, with the data now accessible for further analysis,” states Kwanghun Chung, senior author and associate professor in MIT’s departments of Chemical Engineering and Brain and Cognitive Sciences, as well as a member of The Picower Institute for Learning and Memory. “This technology pipeline paves the way for potentially mapping human brains in their entirety.”
Although this research does not provide an exhaustive map of the entire human brain, it demonstrates an innovative suite of three technologies that enable extensive neuroscience investigations. The study serves as a “proof of concept,” offering numerous examples made possible by this advanced pipeline, such as panoramic views of thousands of neurons within brain regions, intricate cellular landscapes, and detailed subcellular structures interspersed with extracellular molecules. Additionally, the team presents a rich array of quantitative comparisons focused on regions within the Alzheimer’s and non-Alzheimer’s hemispheres.
The capacity to image whole brain hemispheres intact, down to individual synapses — the minute connections facilitating neural circuits — holds significant implications for understanding brain health and disease, according to Chung.
Enhanced Sample Integrity
By using the same brain for various studies, researchers can conduct integrated explorations without the need to piece together disparate data from different brains, which often introduces variability and complicates analysis. A standout feature of the new technology pipeline is that the imaging process does not damage the tissue; rather, it enhances the samples, making them durable and capable of being relabeled multiple times for new research inquiries. Chung’s team successfully employed 20 different antibody labels to highlight various cells and proteins, with plans to expand this number to over a hundred.
“Viewing all functional components — including cell morphology, connectivity, subcellular architectures, and individual synaptic connections within the same brain — is crucial given the substantial variabilities among human brains and the rarity of brain samples,” Chung explains. “This technology enables us to extract vital features from the same brain holistically.”
Moreover, the high scalability and efficiency of the pipeline, where a complete brain hemisphere can be imaged in just 100 hours instead of months, allows for the creation of representative samples across different demographics, such as sex, age, and disease states. Chung envisions establishing a brain bank of fully imaged brains for researchers to explore and re-label as needed.
Three Pioneering Innovations
Chung faced the challenge of assembling a talented team at MIT to develop three key innovations described in the study. Ji Wang, a mechanical engineer and former postdoctoral researcher, created the “Megatome,” a device capable of slicing human brain hemispheres with precision to avoid damage. Juhyuk Park, a materials engineer and former postdoc, engineered the mELAST technology, which enables brain slices to be made clear, flexible, durable, and easily labeled. Meanwhile, Webster Guan, a former MIT chemical engineering graduate student, developed a computational system called “UNSLICE” that reconstructs each hemisphere in 3D, accurately aligning individual blood vessels and neural axons.
Currently, no methodology allows for whole human brain imaging at a subcellular resolution without slicing due to the brain’s significant thickness (3,000 times larger than a mouse brain) and opacity. However, the innovative design of the Megatome by Wang, now at LifeCanvas Technologies, features a blade that vibrates side-to-side quickly, providing wider and more precise cuts than prior slicers, ensuring anatomical integrity and reducing imaging time.
Park’s mELAST technology involves a hydrogel that permeates brain samples, making them optically clear while preserving structural integrity. The combination of this technology with advanced chemical engineering methods enables uniform antibody labeling for high-throughput analysis. Using a customized light sheet microscope, researchers can capture detailed images of whole hemispheres down to individual synapses in about 100 hours.
After imaging, the processed slabs must be computationally reassembled into a coherent picture of the hemisphere. Guan’s UNSLICE employs algorithms to trace blood vessels and align layers, adding to the precision of the reconstructions. A clever labeling strategy allows for effective layer matching based on neural axon tracings.
The paper highlights various achievements, demonstrating how the imaging process allows for comprehensive labeling of entire hemispheres, zooming from large-scale brain structures to individual cells and even subcellular components like synapses. Additional visuals illustrate the diversity of labeled structures, highlighting long axonal connections and variations in cell types such as neurons, astrocytes, and microglia.
Insights into Alzheimer’s Disease
Chung has long collaborated with co-author Matthew Frosch, an Alzheimer’s researcher and director of the brain bank at Massachusetts General Hospital, to study and understand brains impacted by Alzheimer’s disease. With the established pipeline, they began an exploratory investigation, identifying areas with significant neuronal loss in the disease sample compared to the control, subsequently delving deeper into the observed anomalies.
“We approached this project without a rigid experimental framework, simply stating, ‘Let’s look at this slab and see what we can uncover,’” Chung recalls. “We identified specific brain regions with pronounced neuronal loss and expanded our analyses from there.”
“This imaging pipeline grants us nearly limitless access to the biological tissue,” Chung adds. “We can revisit samples to investigate new questions.”
In their investigation, the team concentrated on regions within the orbitofrontal cortex of each hemisphere. They discovered that synapse loss was most pronounced in areas where there was direct overlap with amyloid plaques. Conversely, areas outside of plaque regions exhibited synapse density comparable to that of the non-Alzheimer’s brain.
While drawing conclusions about Alzheimer’s disease from only two samples is premature, this study underscores the capacity to deeply analyze entire human brain hemispheres, setting the stage for future research in this essential area.
This technology not only applies to brain research but can also be leveraged for various other tissues throughout the body.
“We believe this scalable technology platform will enhance our understanding of organ functions and disease mechanisms, ultimately driving the development of new therapies,” the study authors conclude.
In addition to Chung and Frosch, the paper’s contributors include Ji Wang, Juhyuk Park, Webster Guan, Lars A. Gjesteby, Dylan Pollack, Lee Kamentsky, Nicholas B. Evans, Jeff Stirman, Xinyi Gu, Chuanxi Zhao, Slayton Marx, Minyoung E. Kim, Seo Woo Choi, Michael Snyder, David Chavez, Clover Su-Arcaro, Yuxuan Tian, Chang Sin Park, Qiangge Zhang, Dae Hee Yun, Mira Moukheiber, Guoping Feng, X. William Yang, C. Dirk Keene, Patrick R. Hof, Satrajit S. Ghosh, and Laura J. Brattain.
The research received significant funding from the National Institutes of Health, The Picower Institute for Learning and Memory, The JPB Foundation, and the NCSOFT Cultural Foundation.
Photo credit & article inspired by: Massachusetts Institute of Technology