In late 2023, the U.S. Federal Drug Administration approved the first drug that shows promise in slowing the progression of Alzheimer’s disease. This breakthrough is vital, as Alzheimer’s is among several debilitating neurological disorders affecting approximately one-eighth of the global population. While this new treatment marks progress, scientists continue to grapple with a comprehensive understanding of Alzheimer’s and similar conditions.
Lars Gjesteby, a technical staff member and algorithm developer at MIT Lincoln Laboratory’s Human Health and Performance Systems Group, states, “One of the most significant challenges in neuroscience is reconstructing the complex functioning of the human brain at the cellular level. High-resolution, networked brain atlases are essential for enhancing our understanding of brain disorders by identifying contrasts between healthy and diseased brains. Nevertheless, progress has been stalled due to inadequate tools for visualizing and processing vast brain imaging datasets.”
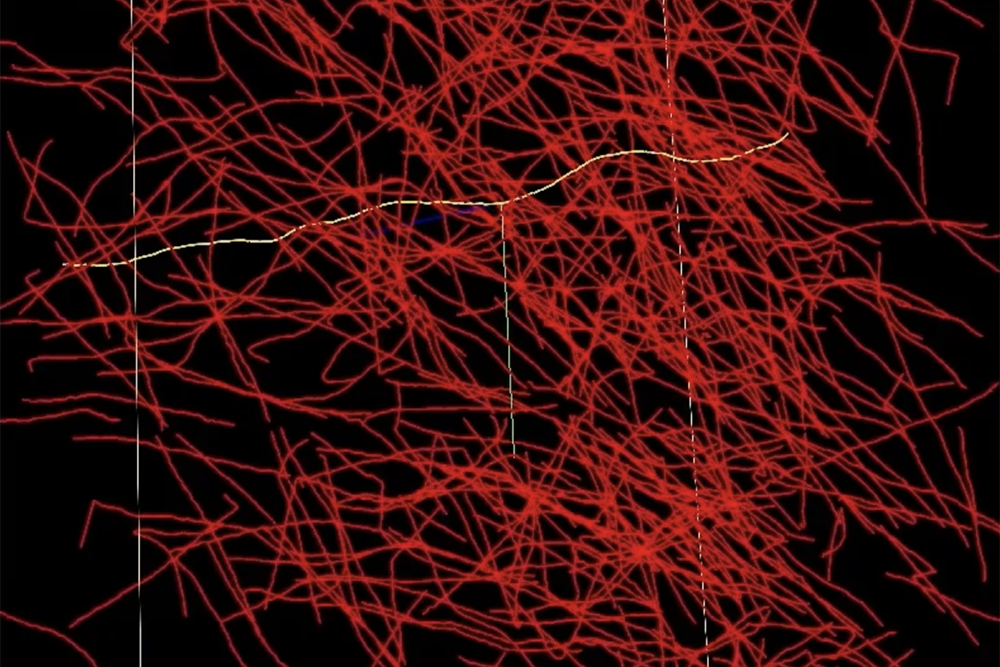
Essentially, a networked brain atlas serves as a comprehensive map of the brain that links structural details to neural functions. Constructing these atlases requires extensive processing and annotation of brain imaging data. For instance, researchers need to trace, measure, and label each axon—those thin fibers connecting neurons. Unfortunately, current methods, including desktop software and manual tools, lack the capacity to manage datasets on the scale of human brains. Consequently, researchers often find themselves overwhelmed by raw data.
Leading the charge is Gjesteby, who is developing the Neuron Tracing and Active Learning Environment (NeuroTrALE). This software pipeline integrates machine learning and supercomputing, simplifying the brain mapping endeavor. NeuroTrALE automates much of the data handling and features an interactive interface, enabling researchers to edit, manipulate, and search for specific patterns easily.
Untangling a Ball of Yarn
One of the standout features of NeuroTrALE is its use of an advanced machine-learning method known as active learning. The algorithms within NeuroTrALE are trained to automatically label incoming data based on established brain imaging datasets. However, when faced with new data, the potential for errors can arise. Active learning empowers users to manually correct these errors, allowing the algorithm to learn and perform better with similar data in the future. This synergy of automation and manual intervention results in accurate data processing, while significantly reducing the user’s workload.
Michael Snyder from the laboratory’s Homeland Decision Support Systems Group elaborates, “Imagine taking an X-ray of a ball of yarn. You see all these overlapping lines. When two lines cross, does it signify that one strand bends at a right angle, or is one going straight up while the other stretches across? Using NeuroTrALE’s active learning, users can trace these strands a few times, training the system to follow them appropriately in future instances. Without NeuroTrALE, researchers would need to trace the axons repeatedly every time.” Snyder works alongside software developer David Chavez on the NeuroTrALE team.
NeuroTrALE alleviates much of the labeling pressure on researchers, enabling faster data processing. Furthermore, its axon tracing algorithms utilize parallel computing, distributing computations across multiple GPUs simultaneously, which enhances processing speed and scalability. The team showcased a remarkable 90% reduction in computing time necessary to process 32 gigabytes of data compared to traditional AI methods.
Significantly, increasing the volume of data does not directly correlate with a similar rise in processing time. Recent research highlighted that a staggering 10,000% increase in dataset size resulted in only 9% and 22% increases in total processing time, respectively, when using two different types of CPUs.
“With approximately 86 billion neurons forming 100 trillion connections within the human brain, the task of manually labeling all neurons in a single brain would take multiple lifetimes,” notes Benjamin Roop, one of the project’s algorithm developers. “This tool can automate the creation of connectomes not just for one individual but for many, paving the way for large-scale studies on brain diseases.”
The Open-Source Road to Discovery
The NeuroTrALE initiative originated as a collaboration between Lincoln Laboratory and Professor Kwanghun Chung’s lab at MIT. The Lincoln Lab team sought to devise a system for Chung Lab researchers to effectively analyze and extract useful insights from their substantial brain imaging data collected via the MIT SuperCloud—a high-performing supercomputer managed by Lincoln Laboratory to support the research community at MIT. Benefiting from its expertise in high-performance computing, image processing, and artificial intelligence, Lincoln Lab is well-equipped to address this challenge.
In 2020, the team made NeuroTrALE available on the SuperCloud, and by 2022, Chung Lab was already yielding results. In one notable study published in Science, researchers employed NeuroTrALE to quantify prefrontal cortex cell density in relation to Alzheimer’s disease, revealing that affected brains exhibited lower cell density in specific areas compared to healthy brains. The same research team also identified regions where harmful neurofibers tend to tangle in the brain tissue of Alzheimer’s patients.
Continuing support from Lincoln Laboratory and funding from the National Institutes of Health (NIH) have facilitated ongoing development of NeuroTrALE. Currently, its user interface tools are being integrated with Google’s Neuroglancer—an open-source, web-based application for visualizing neuroscience data. NeuroTrALE enhances this integration by allowing users to dynamically visualize and edit their annotated data collaboratively, supporting multiple users working on the same data set simultaneously. It also enables the creation and modification of various shapes, such as polygons, points, and lines, to streamline annotation tasks while allowing customization of color displays for dense neuron regions.
“NeuroTrALE offers a platform-agnostic, end-to-end solution that can be swiftly and easily deployed across standalone, virtual, cloud, and high-performance computing environments via containers,” states Adam Michaleas from the laboratory’s Artificial Intelligence Technology Group. “Moreover, it significantly enhances user experience by providing real-time collaboration capabilities within the neuroscience community through data visualization and simultaneous content review.”
Aligned with the NIH’s mission of promoting research sharing, the ultimate goal is to make NeuroTrALE a fully open-source tool accessible to all. Gjesteby emphasizes, “This type of tool is essential for our collective aim to map the entire human brain for research and drug development, fostering a grassroots effort where data and algorithms are open to community access.”
The codebases for the axon tracing, data management, and interactive user interface of NeuroTrALE are publicly available under open-source licenses. Please contact Lars Gjesteby for more information on utilizing NeuroTrALE.
Photo credit & article inspired by: Massachusetts Institute of Technology