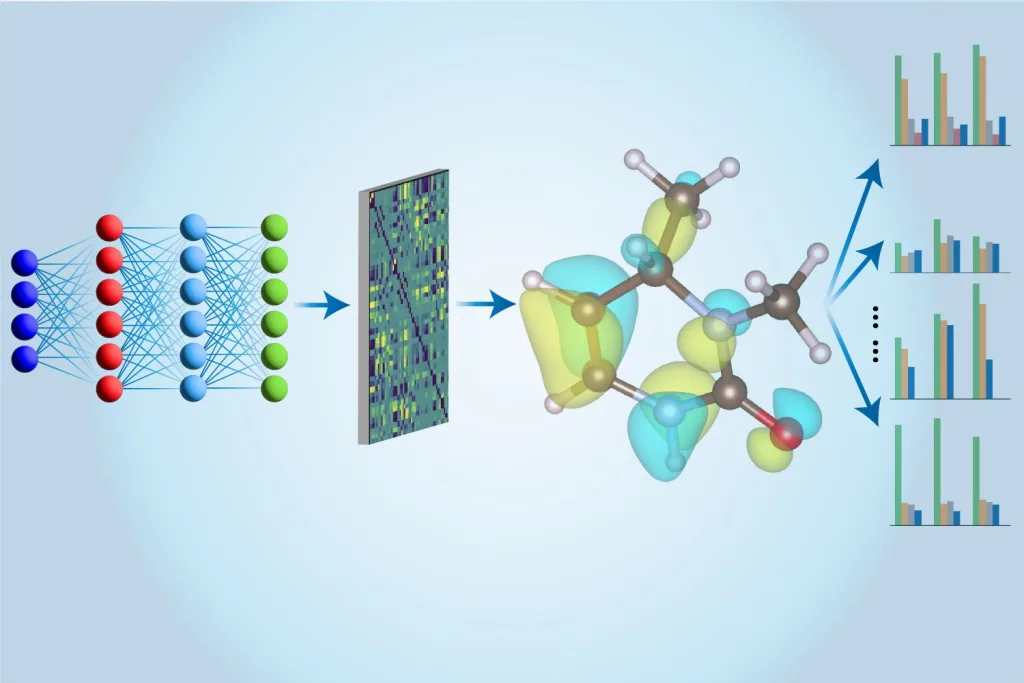
Studying gene expression changes provides invaluable insights into cellular functioning, aiding researchers in comprehending the intricacies of disease development. With approximately 20,000 genes in humans that influence one another through complex interactions, pinpointing which gene groups to target presents a monumental challenge. Furthermore, genes operate collaboratively in regulatory modules, adding another layer of complexity.
Recently, researchers from MIT have laid down groundbreaking theoretical foundations for innovative methods that identify optimal ways to cluster genes into related groups. This technique enables a more efficient understanding of the underlying cause-and-effect dynamics between multiple genes.
A significant advantage of this method is its reliance solely on observational data, negating the need for costly and sometimes impractical interventional experiments. In the long run, this technique could prove beneficial in identifying potential gene targets to influence desired behaviors, paving the way for more precise and tailored treatments for patients.
“Understanding the mechanisms behind cellular states is crucial in genomics. Given the multiscale structure of cells, the manner in which data is summarized can profoundly impact its interpretability and utility,” explains Jiaqi Zhang, a graduate student and co-lead author of the research. He is joined by Ryan Welch, a co-lead author pursuing his master’s degree in engineering, and senior author Caroline Uhler, a professor in the Department of Electrical Engineering and Computer Science (EECS) at MIT.
Observational Data: A New Frontier
The MIT team focused on untangling gene programs; these programs illustrate how genes collaborate to regulate each other during vital biological processes, including cell development and differentiation. Given that studying all 20,000 genes simultaneously is untenable, they employed a technique called causal disentanglement to group related genes into a coherent representation, facilitating efficient exploration of cause-and-effect relationships.
The researchers had previously showcased this technique’s effectiveness with interventional data obtained through variable perturbation. However, accessing such experimental data is often not practical, ethical, or technologically feasible. Relying on observational data alone impedes researchers from comparing gene interactions pre- and post-intervention.
“Most causal disentanglement research presumes the availability of interventions, leaving uncertainty about what can be achieved using solely observational data,” remarks Zhang.
The researchers developed a broader approach, leveraging machine learning algorithms to cluster observed variables (like genes) using only observational data, enabling them to identify causal modules and construct a precise representation of causal mechanisms.
A Layered Approach to Understanding Gene Networks
Using advanced statistical methods, the team computed a mathematical construct known as the variance for the Jacobian of each variable’s score. In this context, causal variables that don’t influence subsequent variables exhibit a variance of zero. They reconstructed the representation in a layered manner, removing bottom-layer variables exhibiting zero variance and iterating backward to determine which gene groups remain interlinked.
“Identifying the variables with zero variance quickly evolves into a complex combinatorial challenge, making the development of an efficient solving algorithm a significant hurdle,” Zhang explains.
Ultimately, their method results in an abstracted representation of the observational data, showcasing interconnected variables that concisely depict the underlying cause-and-effect structure. Each variable encapsulates an aggregated gene group, and the interactions among these variables illustrate regulatory relationships between the groups.
With their theoretical framework validated, the researchers conducted simulations confirming their algorithm’s ability to effectively disentangle meaningful causal representations utilizing observational data alone.
Looking ahead, the researchers aspire to apply their method to real-world genetic data, exploring how it can yield further insights, especially in scenarios where some interventional data might be available. This breakthrough could streamline the process of determining which genes collaborate in specific programs, ultimately leading to the identification of drugs to target those genes for treating various diseases.
Photo credit & article inspired by: Massachusetts Institute of Technology